Biocatalysis
Biocatalysis
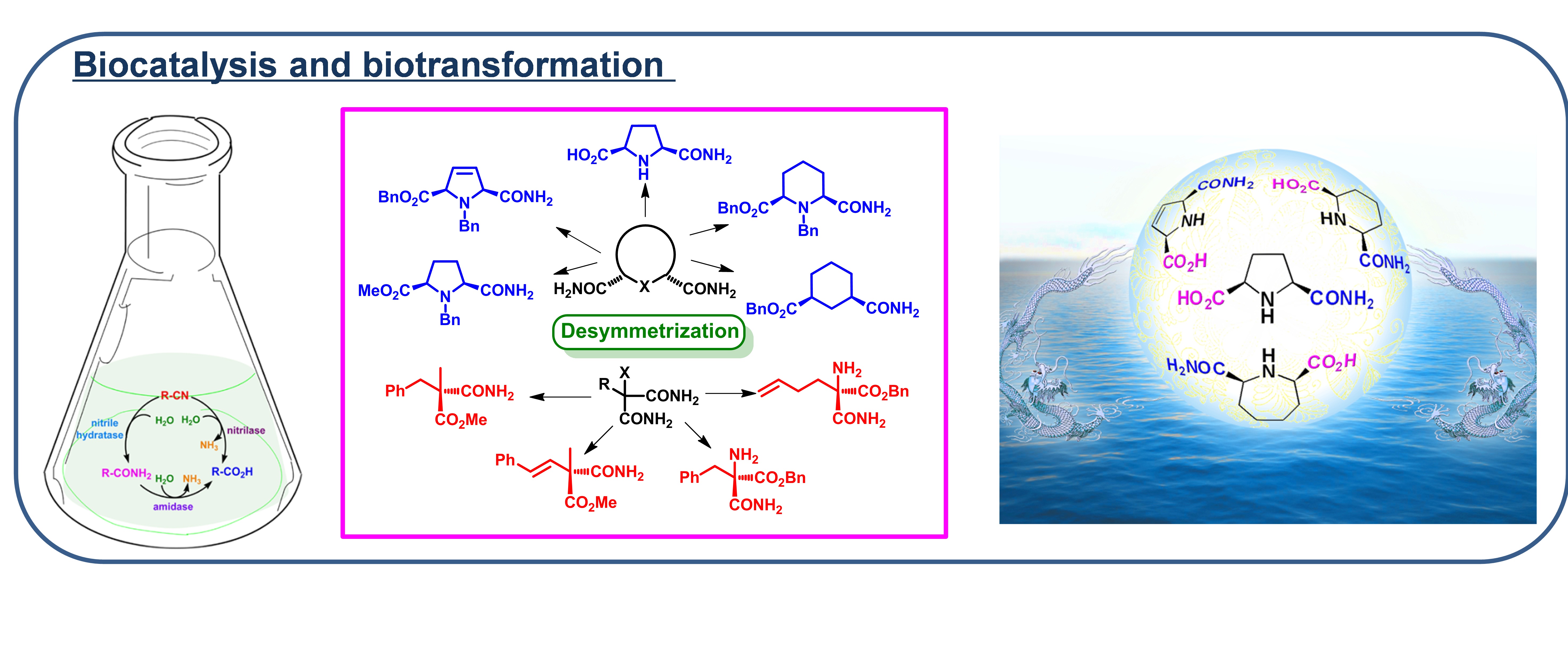
Biotransformations have become an important method in organic synthesis owing to their high efficiency, excellent selectivity and mild reaction conditions. Our group has focused on the biotransformations of racemic nitriles and amides catalyzed by Rhodococcus erythropolis AJ270 for the synthesisi of chiral carboxylic acids and amides, and the mechanism of kinetic resolution of racemic nitriles and amides has also been explored. Now we will design and synthesize novel prochiral and meso-dinitriles and diamides, and develop their biotransformations under whole cell catalysis to synthesize highly enantiopure carboxylic acids and derivatives. The effects of electronic nature and steric hindrance of the substitutents on efficiency and enantioselectivity of the biotransformations will be systematically investigated to find the rules of biocatalytic desymmetrization. On the base of the obtained gene sequence of the amidase, computational method will be applied to construct the protein domain, active center and enzyme-substrate interactions, in order to explore the enantioselective mechanism. Taking the enantiopure carboxylic acids and derivatives as the key starting materials, high efficient transformations will be developed to explore their applications in the synthesis of natural products and bioactive compounds.
With the advent of the AI era, artificial intelligence is influencing various aspects of scientific research. Machine learning methods, which can construct predictive models using limited data, hold the potential to fundamentally transform the traditional trial-and-error research paradigm. We aim to establish quantitative relationships between the structures of enzyme mutants, substrate structures, and their catalytic performance. Such machine learning models could not only guide the design of biosynthetic pathways and mutant selection for targeted products but also provide new insights into reaction mechanisms. However, constructing these models requires a substantial amount of data, which is often limited. To enhance the accuracy of models built with limited data, we focus on addressing two critical challenges: (1) how to collect high-quality and diverse experimental data, and (2) how to design descriptors that precisely characterize the reaction system. By increasing data volume, broadening data scope, and enhancing descriptor precision, we aim to gradually expand the applicability of these models, ultimately achieving AI-assisted rational enzyme engineering.